DeepMind claims early progress in AI-based predictive protein modelling
Google - possessed AI expert, DeepMind, has asserted a "critical achievement" in having the capacity to exhibit the value of man-made brainpower to help with the mind boggling errand of anticipating 3D structures of proteins dependent on their hereditary grouping.
Understanding protein structures is critical in infection analysis and treatment, and could enhance researchers' comprehension of the human body — and possibly supporting protein plan and bioengineering.
Writing in a blog entry about the undertaking to utilize AI to anticipate how proteins crease — now two years in — it states: "The 3D models of proteins that AlphaFold [DeepMind's AI] creates are unquestionably more precise than any that have preceded — gaining huge ground on one of the center difficulties in science."
There are different logical techniques for foreseeing the local 3D condition of protein atoms (i.e. how the protein bind folds to touch base at the local state) from remaining amino acids in DNA.
Be that as it may, displaying the 3D structure is an exceedingly perplexing errand, given what number of changes there can be by virtue of protein collapsing being subject to variables, for example, collaborations between amino acids.
There's even a publicly supported amusement (FoldIt) that attempts to use human instinct to anticipate useful protein shapes.
DeepMind says its methodology heaps of earlier research in utilizing huge information to attempt to anticipate protein structures.
Explicitly it's applying profound learning ways to deal with genomic information.
"Luckily, the field of genomics is very wealthy in information because of the quick decrease in the expense of hereditary sequencing. Subsequently, profound learning ways to deal with the forecast issue that depend on genomic information have turned out to be progressively well known over the most recent couple of years. DeepMind's work on this issue brought about AlphaFold, which we submitted to CASP [Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction] this year," it writes in the blog entry.
"We're pleased to be a piece of what the CASP coordinators have called "extraordinary advancement in the capacity of computational strategies to anticipate protein structure," setting first in rankings among the groups that entered (our entrance is A7D)."
"Our group concentrated explicitly on the difficult issue of demonstrating target shapes sans preparation, without utilizing recently tackled proteins as layouts. We accomplished a high level of exactness while foreseeing the physical properties of a protein structure, and after that utilized two unmistakable techniques to develop forecasts of full protein structures," it includes.
DeepMind says the two techniques it utilized depended on utilizing profound neural systems prepared to anticipate protein properties from its hereditary arrangement.
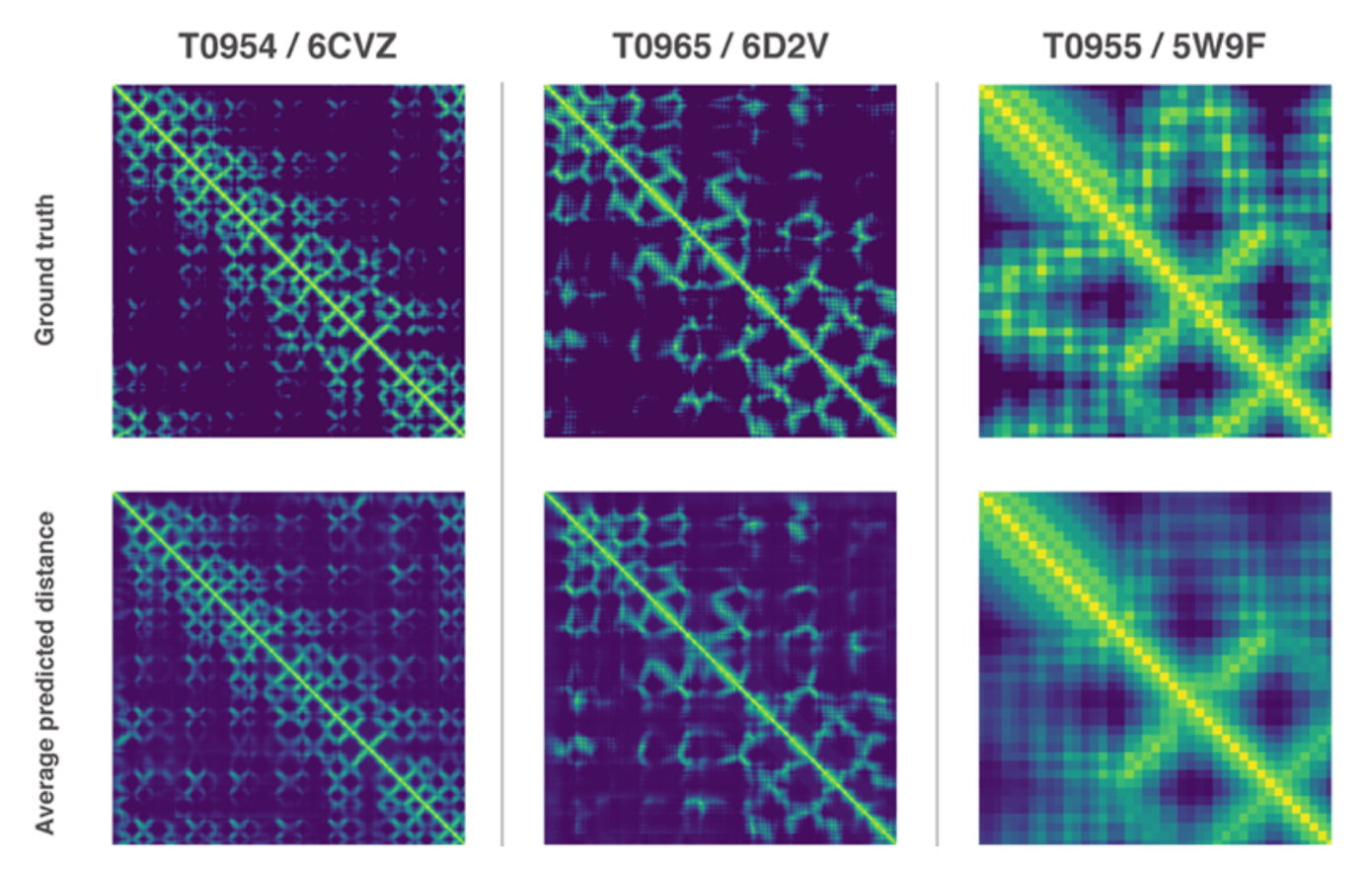
"The properties our systems foresee are: (a) the separations between sets of amino acids and (b) the edges between synthetic bonds that associate those amino acids. The primary improvement is a development on usually utilized methods that gauge whether sets of amino acids are close to one another," it clarifies.
"We prepared a neural system to anticipate a different conveyance of separations between each match of deposits in a protein. These probabilities were then joined into a score that gauges how precise a proposed protein structure is. We additionally prepared a different neural system that utilizes all separations in total to assess how shut the proposed structure is to the correct answer."
It at that point utilized new strategies to endeavor to build expectations of protein structures, seeking known structures that coordinated its forecasts.
"Our first strategy based on methods regularly utilized in basic science, and more than once supplanted bits of a protein structure with new protein parts. We prepared a generative neural system to design new parts, which were utilized to constantly enhance the score of the proposed protein structure," it composes.
"The second strategy upgraded scores through angle drop — a scientific procedure normally utilized in machine learning for making little, gradual enhancements — which brought about exceedingly exact structures. This procedure was connected to whole protein affixes as opposed to pieces that must be collapsed independently before being amassed, diminishing the multifaceted nature of the forecast procedure."
DeepMind portrays the outcomes accomplished up to this point as "early indications of advancement in protein collapsing" utilizing computational techniques — asserting they illustrate "the utility of AI for logical revelation".
Despite the fact that it likewise underscores it's still early days for the profound learning approach having any sort of "quantifiable effect".
"Despite the fact that there's much more work to do before we're ready to quantifiably affect treating infections, dealing with the earth, and the sky is the limit from there, we realize the potential is tremendous," it composes. "With a committed group concentrated on diving into how machine learning can propel the universe of science, we're anticipating seeing the numerous ways our innovation can have any kind of effect."
Understanding protein structures is critical in infection analysis and treatment, and could enhance researchers' comprehension of the human body — and possibly supporting protein plan and bioengineering.
Writing in a blog entry about the undertaking to utilize AI to anticipate how proteins crease — now two years in — it states: "The 3D models of proteins that AlphaFold [DeepMind's AI] creates are unquestionably more precise than any that have preceded — gaining huge ground on one of the center difficulties in science."
There are different logical techniques for foreseeing the local 3D condition of protein atoms (i.e. how the protein bind folds to touch base at the local state) from remaining amino acids in DNA.
Be that as it may, displaying the 3D structure is an exceedingly perplexing errand, given what number of changes there can be by virtue of protein collapsing being subject to variables, for example, collaborations between amino acids.
There's even a publicly supported amusement (FoldIt) that attempts to use human instinct to anticipate useful protein shapes.
DeepMind says its methodology heaps of earlier research in utilizing huge information to attempt to anticipate protein structures.
Explicitly it's applying profound learning ways to deal with genomic information.
"Luckily, the field of genomics is very wealthy in information because of the quick decrease in the expense of hereditary sequencing. Subsequently, profound learning ways to deal with the forecast issue that depend on genomic information have turned out to be progressively well known over the most recent couple of years. DeepMind's work on this issue brought about AlphaFold, which we submitted to CASP [Community Wide Experiment on the Critical Assessment of Techniques for Protein Structure Prediction] this year," it writes in the blog entry.
"We're pleased to be a piece of what the CASP coordinators have called "extraordinary advancement in the capacity of computational strategies to anticipate protein structure," setting first in rankings among the groups that entered (our entrance is A7D)."
"Our group concentrated explicitly on the difficult issue of demonstrating target shapes sans preparation, without utilizing recently tackled proteins as layouts. We accomplished a high level of exactness while foreseeing the physical properties of a protein structure, and after that utilized two unmistakable techniques to develop forecasts of full protein structures," it includes.
DeepMind says the two techniques it utilized depended on utilizing profound neural systems prepared to anticipate protein properties from its hereditary arrangement.
"The properties our systems foresee are: (a) the separations between sets of amino acids and (b) the edges between synthetic bonds that associate those amino acids. The primary improvement is a development on usually utilized methods that gauge whether sets of amino acids are close to one another," it clarifies.
"We prepared a neural system to anticipate a different conveyance of separations between each match of deposits in a protein. These probabilities were then joined into a score that gauges how precise a proposed protein structure is. We additionally prepared a different neural system that utilizes all separations in total to assess how shut the proposed structure is to the correct answer."
It at that point utilized new strategies to endeavor to build expectations of protein structures, seeking known structures that coordinated its forecasts.
"Our first strategy based on methods regularly utilized in basic science, and more than once supplanted bits of a protein structure with new protein parts. We prepared a generative neural system to design new parts, which were utilized to constantly enhance the score of the proposed protein structure," it composes.
"The second strategy upgraded scores through angle drop — a scientific procedure normally utilized in machine learning for making little, gradual enhancements — which brought about exceedingly exact structures. This procedure was connected to whole protein affixes as opposed to pieces that must be collapsed independently before being amassed, diminishing the multifaceted nature of the forecast procedure."
DeepMind portrays the outcomes accomplished up to this point as "early indications of advancement in protein collapsing" utilizing computational techniques — asserting they illustrate "the utility of AI for logical revelation".
Despite the fact that it likewise underscores it's still early days for the profound learning approach having any sort of "quantifiable effect".
"Despite the fact that there's much more work to do before we're ready to quantifiably affect treating infections, dealing with the earth, and the sky is the limit from there, we realize the potential is tremendous," it composes. "With a committed group concentrated on diving into how machine learning can propel the universe of science, we're anticipating seeing the numerous ways our innovation can have any kind of effect."
DeepMind claims early progress in AI-based predictive protein modelling
![]() Reviewed by Tayyab Tahir
on
03:47
Rating:
Reviewed by Tayyab Tahir
on
03:47
Rating:
No comments: